######### STEP 2: CREATE YOUR OBJECT #########
#take your individual section or combined dataset and turn it into an mFISH object for analysis
myobj <- ruMake(mydata)[1] "Creating object..."Kaitlin Sullivan
The following video tutorial demonstrates the structure of the formal mFISH class object.
Read below for code and a visual representation of the mFISH object’s structure.
mFISH Object with ruMake()mFISH Object Structure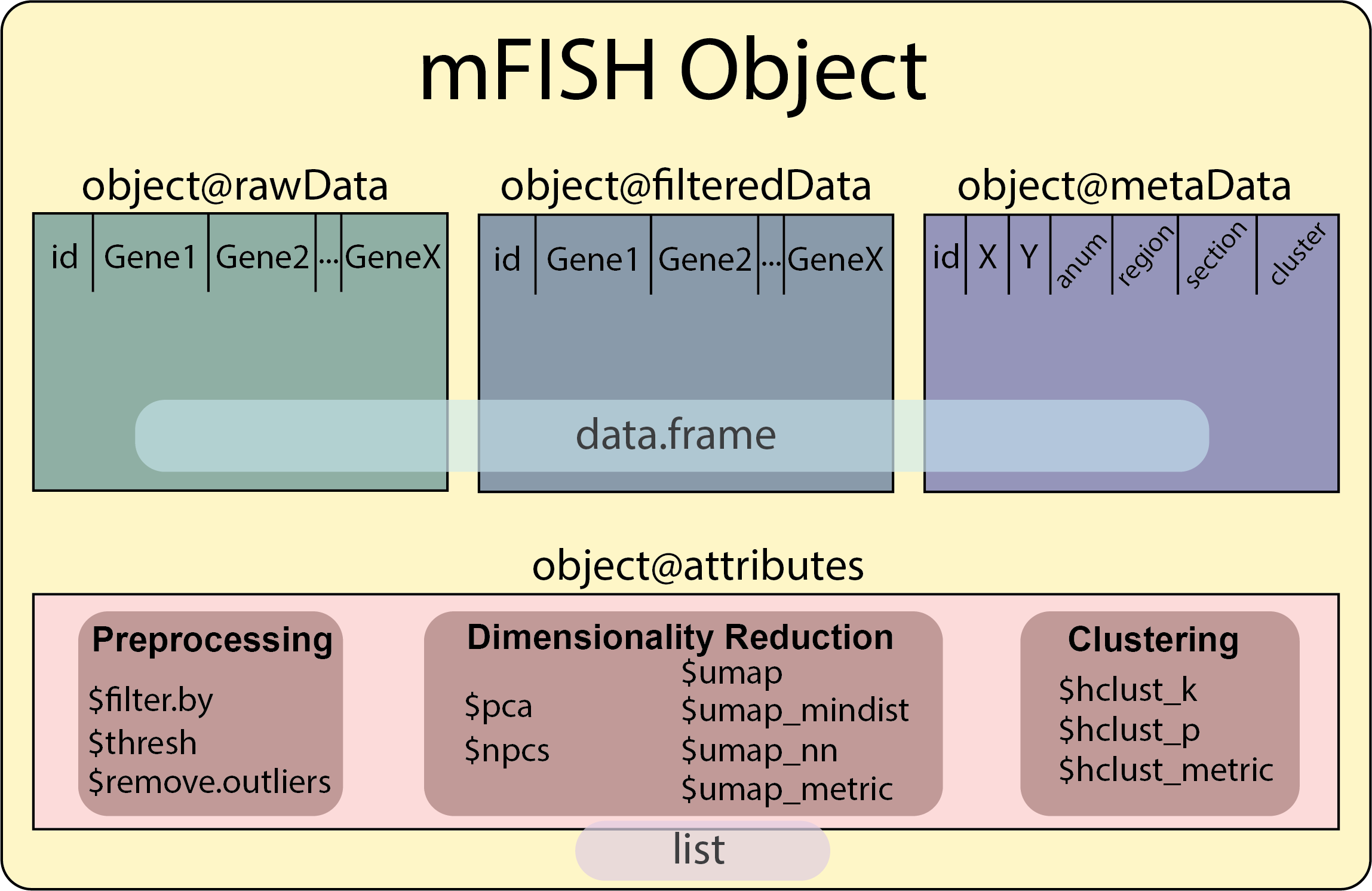
The mFISH object contains four slots:
@rawDataThis contains the original data frame, without metadata, from ruRead().
Nnat Synpr Pcp4 Slc17a7 Cdh9 Ctgf Slc17a6 Lxn
1 0.1834896 0.0000000 5.5177946 0.0000000 0.7358308 0.3679154 0.73583077 0
2 0.0000000 0.0000000 0.0000000 0.0000000 2.1668655 0.0000000 2.16686552 0
3 0.0000000 0.0000000 0.0000000 0.6694704 0.0000000 0.0000000 1.00420555 0
4 0.0000000 0.2438931 0.4888655 13.1972094 0.2438931 0.4888655 0.73275862 0
5 0.1613582 0.2426580 0.8893321 38.7961045 0.3233371 0.3233371 0.08067912 0
6 0.0000000 12.5748426 0.0000000 23.0532699 0.0000000 0.0000000 0.00000000 0
Slc30a3 Gfra1 Spon1 Gnb4 RSC LEC id
1 0.0000000 0.0000000 1.1037462 0.0000000 0 0 1
2 0.7216701 0.0000000 0.0000000 0.0000000 0 0 2
3 0.0000000 0.3347352 0.6694704 0.6694704 0 0 3
4 0.0000000 0.0000000 0.0000000 0.0000000 0 0 4
5 0.4040162 0.0000000 3.1520710 2.0207016 0 0 5
6 0.0000000 1.3976102 0.0000000 0.0000000 0 0 6@filteredDataThis contains a data frame of the normalized and filtered data from downstream analysis.
For now this is empty, however this slot will be populated automatically during analysis with the goFISH Shiny app or generated via ruFilter() and ruProcess()).
@metaDataThis contains a data frame of all the metadata from the initial ruRead() data frame.
Nnat Synpr Pcp4 Slc17a7 Cdh9 Ctgf Slc17a6 Lxn
1 0.1834896 0.0000000 5.5177946 0.0000000 0.7358308 0.3679154 0.73583077 0
2 0.0000000 0.0000000 0.0000000 0.0000000 2.1668655 0.0000000 2.16686552 0
3 0.0000000 0.0000000 0.0000000 0.6694704 0.0000000 0.0000000 1.00420555 0
4 0.0000000 0.2438931 0.4888655 13.1972094 0.2438931 0.4888655 0.73275862 0
5 0.1613582 0.2426580 0.8893321 38.7961045 0.3233371 0.3233371 0.08067912 0
6 0.0000000 12.5748426 0.0000000 23.0532699 0.0000000 0.0000000 0.00000000 0
Slc30a3 Gfra1 Spon1 Gnb4 RSC LEC id
1 0.0000000 0.0000000 1.1037462 0.0000000 0 0 1
2 0.7216701 0.0000000 0.0000000 0.0000000 0 0 2
3 0.0000000 0.3347352 0.6694704 0.6694704 0 0 3
4 0.0000000 0.0000000 0.0000000 0.0000000 0 0 4
5 0.4040162 0.0000000 3.1520710 2.0207016 0 0 5
6 0.0000000 1.3976102 0.0000000 0.0000000 0 0 6@attributesThis contains a list of all the important variables used throughout analysis to assist with code reproducibility.
$filter.by
[1] NA
$thresh
[1] NA
$umap_nn
[1] NA
$umap_mindist
[1] NA
$umap_metric
[1] NA
$hclust_k
[1] NA